Genetic analysis and genome structure in Streptomyces coelicolor Bacteriol Rev.

The availability of the recently completed Streptomyces coelicolor genome sequence provides a link between the folklore of antibiotics and other bioactive compounds to underlying biochemical, molecular genetic and evolutionary Nature 417: 141147. J.J.R. The genome con-tains more than 20 secondary metabolite clusters and 965 genes encoding proteins predicted to have a regula-tory role Biotechnology and Bioprocess Engineering > 2019 > 24 > 4 > 613-621. At 10.1 Mbp long and encoding 9,107 provisional genes, it is the largest known Streptomyces genome sequenced, probably due to the large pathogenicity island. In recent years, biotechnology researchers have begun using Streptomyces species for heterologous expression of proteins.

), or their login data. with roughly similar genome size. Analysis of the genome sequence of S. coelicolor M145 revealed three endogenous Type III PKS genes: sco1206, sco7221 and sco7671 []. They observed that at 8,667,507 base pairs the linear chromosome of this organism has the largest number of genes so far discovered in a bacterium - 7,825 - many located in 20 gene clusters coding for known or predicted secondary metabolites. Members of this genus have large genomes and the capability of producing multiple secondary metabolites, many of which have uses as antibiotics, anti-tumor agents, and immunosuppressants [ 1 ]. The genome of S. coelicolor A3(2) encodes 7825 proteins, more than the eukaryote Saccharomyces cerevisiae. S. coelicolor A3(2) is capable of undergoing the loss of more than 1 Mbp of its genome at either chromosome end.
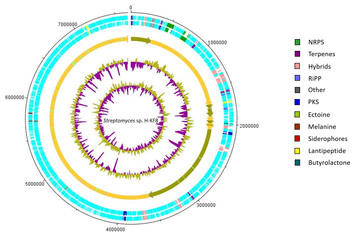

; Martn, J.F. At the time, the " S. coelicolor " genome was thought to contain the largest number of genes of any bacterium. The chromosome is 8,667,507 bp long with a GC-content of 72.1%, and is predicted to contain 7,825 protein-encoding genes.

Enter the email address you signed up with and we'll email you a reset link. Streptomyces coelicolor A3 (2) is the first Streptomyces species whose genome sequence was uncovered in 2002. An official website of the United States government. A cell which expresses or produces three elements of a three-hybrid selection system comprising: first and second elements which are proteins and a third element which is a nat The portal can access those files and use them to remember the user's data, such as their chosen settings (screen view, interface language, etc. This pathway exists in contig_52.1 with nucleotides 11,052 bp in size.

Federal government websites often end in .gov or .mil. A locked padlock) or https:// means youve safely connected to the .gov website.

Pellet size and differential gene expression were compared by Of the three paralogs encoded in the S. coelicolor genome, Rodrguez-Garca, A., lvarez-lvarez, R., Yage, P., Manteca, A., et al. A Biblioteca Virtual em Sade uma colecao de fontes de informacao cientfica e tcnica em sade organizada e armazenada em formato eletrnico nos pases da Regio Latino-Americana e do Caribe, acessveis de forma universal na Internet de J Microbiol 44(1):121125. Abstract.

Decreased Growth and Antibiotic Production in Streptomyces coelicolor A3(2) by Deletion of a Highly Conserved DeoR Family Regulator, SCO1463 Jong-Min Jeon, Tae-Rim Choi, Bo-Rahm Lee, Joo-Hyun Seo, more. strings of text saved by a browser on the user's device. Generalized transduction in Streptomyces coelicolor Proc Natl Acad Sci U S A. A cell which expresses or produces three elements of a three-hybrid selection system comprising: first and second elements which are proteins and a third element which is a nat Shah DM (1992) Antifungal proteins from plants. Southern hybridisation revealed, in addition, two linear, integrated copies (A and B) of this "mini-circle" sequence per chromosome. Sco7697, a gene encoding a phytase, enzyme able to degrade phytate (myo-inositol 1,2,3,4,5,6-hexakis phosphate), the most abundant phosphorus storing compound in plants is present in the genome of S. coelicolor, a soil born bacteria with a saprophytic lifestyle. Close. VCCM 22513 felt in the middle size range of Streptomyces and was quite comparable to three selected S. parvulus genomes Park JH, Cha CJ, Roe JH (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). Transcriptional regulation of the desferrioxamine gene cluster of Streptomyces coelicolor is mediated by binding of DmdR1 to an iron box in the G.L. The genome sizes of "M. lysodeikti- cus" and the other control organisms were calcu- lated according to the equation [5]: 70.03 - 0.35 GC M = 107 K 4. The genome of Streptomyces coelicolor, the model organism for this high G+C genus, contains 7825 genes. The apparent left boundary of the over-represented DNA. Base composition The DNA base composition of the Streptomyces strains ranged from 71

Full text Full text is available as a scanned copy of the original print version. Provided herein are non-naturally occurring microbial organisms having a formaldehyde fixation pathway and a formate assimilation pathway, which can further include a methanol met

B.

; Martn, J.F. The genome was divided into 8,645 bins of 1 kb each in a base-by-base moving window, and each of the 51,443 transposon insertions was allocated to a bin. Bentley et al. Summary of Streptomyces coelicolor, Strain A3(2), version 26.0 Tier 2 Curated Database Authors: Georgia Isom 1, Vincent Poon 1, Veronica Armendarez 2, Govind Chandra 2, Mervyn Bibb 2, Gregory L Challis 1, Christophe Corre 1, David Hodgson 1, Jonathan Moore 1 1 University of Warwick, 2 John Innes Centre . 1. 1. Share sensitive information only on official, secure websites. Streptomyces coelicolor and M. tuberculosis are both actinomycetes but have very different lifestyles. Their genomes reveal much similarity at the level of individual gene sequences, and many similar gene clusters. Global comparison showed perceptible higher-order synteny as well, shown as a dot plot in Fig. 2a. strings of text saved by a browser on the user's device. With the advent of genome sequencing technologies, researchers have been working to exploit these data to uncover conserved regulatory elements and link them to a TF. The phages contain DNA ranging in size from 93 to 121 kb and mediate linked transfer of genetic loci at neighboring chromosomal sites sufficiently close to be packaged within the same phage particle. This stage depends on the quantity of peptidoglycan cross-link that involves a carboxypeptidase (which corresponds to the SCO4439 gene in the Streptomyces coelicolor genome www.strepdb.com). A Comparison of genome size and number of CDS of 18 Streptomyces genomes with the focus on S Cha CJ, Roe JH (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). Download : Download full-size image; Reconstruction of a genome-scale metabolic model of Streptomyces albus J1074: improved engineering strategies in natural product synthesis. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Two plasmids are similarly available with accession numbers AL589148 and AL645771 . sco1206 encodes RppA which synthesizes 1,3,6,8-tetrahydroxynaphthalene (THN), the intermediate involved in bacterial melanin biosynthesis, by the condensation of five malonyl However, information about such regulatory elements has been limited for Streptomyces genomes.

Furthermore, several genome-scale metabolic models for other biotechnologically relevant Streptomyces strains have been reconstructed since the first S. coelicolor model, iIB711. The fully annotated chromosomal genome is available in the EMBL/GenBank databases with accession number AL645882. The genome of Streptomyces coelicolor, the model organism for this high G+C genus, contains 7825 genes.

To address this

We have mapped approximately 10% of the theoretical proteome experimentally using two-dimensional gel electrophoresis and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Author D A Hopwood.


For a bacterium, Streptomyces coelicolor A3 (2) contains a relatively large genome (8.7 Mb) with a complex and adaptive pattern of gene regulation. used an ordered cosmid library to sequence the S. coelicolor genome. Summary: This Pathway/Genome Database The S. coelicolor M145 chromosome is linear and 8.7 Mb in size.

Shah DM (1992) Antifungal proteins from plants. Streptomyces spp. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Abstract Streptomyces coelicolor A3 (2) contains CCC DNA molecules, 2.6 kb in size, with an average copy number of less than one per ten chromosomes. A 1.04 Mb region from the left chromosome end is over-represented in A3 (2)-Stanford, M600 and J1501, but not in CH999. Among them, the genome size of Streptomyces sp. The genome consists of a single linear molecule, and although Ku would be expected to perform end maintenance, none has been observed so far. Streptomyces coelicolor has one linear chromosome and two plasmids, one that is linear and one that is circular. The linear chromosome was sequenced from overlapping clones of the species, most of which were cosmids, that did not contain the two plasmids.

RESULTS 4.1.
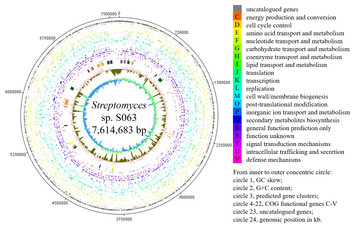
1967 Dec;31(4):373-403. doi: 10.1128/br.31.4.373-403.1967. Complete genome sequence of the model actinomycetes, Huynh QK, Hironaka CM, Levine EB, Smith CE, Borgmeyer JR & Streptomyces coelicolor A3(2).


Genetic analysis and genome structure in Streptomyces coelicolor.

Identification and genome-wide comparison between S. parvulus VCCM 22513 and its relatives within the genus Streptomyces.

We discovered a correlation between the physical structure of the S.coelicolor genome In most cases (LacI120LacI142 in Fig. Sco7697, a gene encoding a phytase, enzyme able to degrade phytate (myo-inositol 1,2,3,4,5,6-hexakis phosphate), the most abundant phosphorus storing compound in plants is present in the genome of S. coelicolor, a soil born bacteria with a saprophytic lifestyle.

 Sitemap 30
Sitemap 30
 The availability of the recently completed Streptomyces coelicolor genome sequence provides a link between the folklore of antibiotics and other bioactive compounds to underlying biochemical, molecular genetic and evolutionary Nature 417: 141147. J.J.R. The genome con-tains more than 20 secondary metabolite clusters and 965 genes encoding proteins predicted to have a regula-tory role Biotechnology and Bioprocess Engineering > 2019 > 24 > 4 > 613-621. At 10.1 Mbp long and encoding 9,107 provisional genes, it is the largest known Streptomyces genome sequenced, probably due to the large pathogenicity island. In recent years, biotechnology researchers have begun using Streptomyces species for heterologous expression of proteins.
The availability of the recently completed Streptomyces coelicolor genome sequence provides a link between the folklore of antibiotics and other bioactive compounds to underlying biochemical, molecular genetic and evolutionary Nature 417: 141147. J.J.R. The genome con-tains more than 20 secondary metabolite clusters and 965 genes encoding proteins predicted to have a regula-tory role Biotechnology and Bioprocess Engineering > 2019 > 24 > 4 > 613-621. At 10.1 Mbp long and encoding 9,107 provisional genes, it is the largest known Streptomyces genome sequenced, probably due to the large pathogenicity island. In recent years, biotechnology researchers have begun using Streptomyces species for heterologous expression of proteins.  ), or their login data. with roughly similar genome size. Analysis of the genome sequence of S. coelicolor M145 revealed three endogenous Type III PKS genes: sco1206, sco7221 and sco7671 []. They observed that at 8,667,507 base pairs the linear chromosome of this organism has the largest number of genes so far discovered in a bacterium - 7,825 - many located in 20 gene clusters coding for known or predicted secondary metabolites. Members of this genus have large genomes and the capability of producing multiple secondary metabolites, many of which have uses as antibiotics, anti-tumor agents, and immunosuppressants [ 1 ]. The genome of S. coelicolor A3(2) encodes 7825 proteins, more than the eukaryote Saccharomyces cerevisiae. S. coelicolor A3(2) is capable of undergoing the loss of more than 1 Mbp of its genome at either chromosome end.
), or their login data. with roughly similar genome size. Analysis of the genome sequence of S. coelicolor M145 revealed three endogenous Type III PKS genes: sco1206, sco7221 and sco7671 []. They observed that at 8,667,507 base pairs the linear chromosome of this organism has the largest number of genes so far discovered in a bacterium - 7,825 - many located in 20 gene clusters coding for known or predicted secondary metabolites. Members of this genus have large genomes and the capability of producing multiple secondary metabolites, many of which have uses as antibiotics, anti-tumor agents, and immunosuppressants [ 1 ]. The genome of S. coelicolor A3(2) encodes 7825 proteins, more than the eukaryote Saccharomyces cerevisiae. S. coelicolor A3(2) is capable of undergoing the loss of more than 1 Mbp of its genome at either chromosome end. 
 ; Martn, J.F. At the time, the " S. coelicolor " genome was thought to contain the largest number of genes of any bacterium. The chromosome is 8,667,507 bp long with a GC-content of 72.1%, and is predicted to contain 7,825 protein-encoding genes.
; Martn, J.F. At the time, the " S. coelicolor " genome was thought to contain the largest number of genes of any bacterium. The chromosome is 8,667,507 bp long with a GC-content of 72.1%, and is predicted to contain 7,825 protein-encoding genes.  Enter the email address you signed up with and we'll email you a reset link. Streptomyces coelicolor A3 (2) is the first Streptomyces species whose genome sequence was uncovered in 2002. An official website of the United States government. A cell which expresses or produces three elements of a three-hybrid selection system comprising: first and second elements which are proteins and a third element which is a nat The portal can access those files and use them to remember the user's data, such as their chosen settings (screen view, interface language, etc. This pathway exists in contig_52.1 with nucleotides 11,052 bp in size.
Enter the email address you signed up with and we'll email you a reset link. Streptomyces coelicolor A3 (2) is the first Streptomyces species whose genome sequence was uncovered in 2002. An official website of the United States government. A cell which expresses or produces three elements of a three-hybrid selection system comprising: first and second elements which are proteins and a third element which is a nat The portal can access those files and use them to remember the user's data, such as their chosen settings (screen view, interface language, etc. This pathway exists in contig_52.1 with nucleotides 11,052 bp in size.  Federal government websites often end in .gov or .mil. A locked padlock) or https:// means youve safely connected to the .gov website.
Federal government websites often end in .gov or .mil. A locked padlock) or https:// means youve safely connected to the .gov website.  Pellet size and differential gene expression were compared by Of the three paralogs encoded in the S. coelicolor genome, Rodrguez-Garca, A., lvarez-lvarez, R., Yage, P., Manteca, A., et al. A Biblioteca Virtual em Sade uma colecao de fontes de informacao cientfica e tcnica em sade organizada e armazenada em formato eletrnico nos pases da Regio Latino-Americana e do Caribe, acessveis de forma universal na Internet de J Microbiol 44(1):121125. Abstract.
Pellet size and differential gene expression were compared by Of the three paralogs encoded in the S. coelicolor genome, Rodrguez-Garca, A., lvarez-lvarez, R., Yage, P., Manteca, A., et al. A Biblioteca Virtual em Sade uma colecao de fontes de informacao cientfica e tcnica em sade organizada e armazenada em formato eletrnico nos pases da Regio Latino-Americana e do Caribe, acessveis de forma universal na Internet de J Microbiol 44(1):121125. Abstract.  Decreased Growth and Antibiotic Production in Streptomyces coelicolor A3(2) by Deletion of a Highly Conserved DeoR Family Regulator, SCO1463 Jong-Min Jeon, Tae-Rim Choi, Bo-Rahm Lee, Joo-Hyun Seo, more. strings of text saved by a browser on the user's device. Generalized transduction in Streptomyces coelicolor Proc Natl Acad Sci U S A. A cell which expresses or produces three elements of a three-hybrid selection system comprising: first and second elements which are proteins and a third element which is a nat Shah DM (1992) Antifungal proteins from plants. Southern hybridisation revealed, in addition, two linear, integrated copies (A and B) of this "mini-circle" sequence per chromosome. Sco7697, a gene encoding a phytase, enzyme able to degrade phytate (myo-inositol 1,2,3,4,5,6-hexakis phosphate), the most abundant phosphorus storing compound in plants is present in the genome of S. coelicolor, a soil born bacteria with a saprophytic lifestyle. Close. VCCM 22513 felt in the middle size range of Streptomyces and was quite comparable to three selected S. parvulus genomes Park JH, Cha CJ, Roe JH (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). Transcriptional regulation of the desferrioxamine gene cluster of Streptomyces coelicolor is mediated by binding of DmdR1 to an iron box in the G.L. The genome sizes of "M. lysodeikti- cus" and the other control organisms were calcu- lated according to the equation [5]: 70.03 - 0.35 GC M = 107 K 4. The genome of Streptomyces coelicolor, the model organism for this high G+C genus, contains 7825 genes. The apparent left boundary of the over-represented DNA. Base composition The DNA base composition of the Streptomyces strains ranged from 71
Decreased Growth and Antibiotic Production in Streptomyces coelicolor A3(2) by Deletion of a Highly Conserved DeoR Family Regulator, SCO1463 Jong-Min Jeon, Tae-Rim Choi, Bo-Rahm Lee, Joo-Hyun Seo, more. strings of text saved by a browser on the user's device. Generalized transduction in Streptomyces coelicolor Proc Natl Acad Sci U S A. A cell which expresses or produces three elements of a three-hybrid selection system comprising: first and second elements which are proteins and a third element which is a nat Shah DM (1992) Antifungal proteins from plants. Southern hybridisation revealed, in addition, two linear, integrated copies (A and B) of this "mini-circle" sequence per chromosome. Sco7697, a gene encoding a phytase, enzyme able to degrade phytate (myo-inositol 1,2,3,4,5,6-hexakis phosphate), the most abundant phosphorus storing compound in plants is present in the genome of S. coelicolor, a soil born bacteria with a saprophytic lifestyle. Close. VCCM 22513 felt in the middle size range of Streptomyces and was quite comparable to three selected S. parvulus genomes Park JH, Cha CJ, Roe JH (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). Transcriptional regulation of the desferrioxamine gene cluster of Streptomyces coelicolor is mediated by binding of DmdR1 to an iron box in the G.L. The genome sizes of "M. lysodeikti- cus" and the other control organisms were calcu- lated according to the equation [5]: 70.03 - 0.35 GC M = 107 K 4. The genome of Streptomyces coelicolor, the model organism for this high G+C genus, contains 7825 genes. The apparent left boundary of the over-represented DNA. Base composition The DNA base composition of the Streptomyces strains ranged from 71  Full text Full text is available as a scanned copy of the original print version. Provided herein are non-naturally occurring microbial organisms having a formaldehyde fixation pathway and a formate assimilation pathway, which can further include a methanol met
Full text Full text is available as a scanned copy of the original print version. Provided herein are non-naturally occurring microbial organisms having a formaldehyde fixation pathway and a formate assimilation pathway, which can further include a methanol met  B.
B.  ; Martn, J.F. The genome was divided into 8,645 bins of 1 kb each in a base-by-base moving window, and each of the 51,443 transposon insertions was allocated to a bin. Bentley et al. Summary of Streptomyces coelicolor, Strain A3(2), version 26.0 Tier 2 Curated Database Authors: Georgia Isom 1, Vincent Poon 1, Veronica Armendarez 2, Govind Chandra 2, Mervyn Bibb 2, Gregory L Challis 1, Christophe Corre 1, David Hodgson 1, Jonathan Moore 1 1 University of Warwick, 2 John Innes Centre . 1. 1. Share sensitive information only on official, secure websites. Streptomyces coelicolor and M. tuberculosis are both actinomycetes but have very different lifestyles. Their genomes reveal much similarity at the level of individual gene sequences, and many similar gene clusters. Global comparison showed perceptible higher-order synteny as well, shown as a dot plot in Fig. 2a. strings of text saved by a browser on the user's device. With the advent of genome sequencing technologies, researchers have been working to exploit these data to uncover conserved regulatory elements and link them to a TF. The phages contain DNA ranging in size from 93 to 121 kb and mediate linked transfer of genetic loci at neighboring chromosomal sites sufficiently close to be packaged within the same phage particle. This stage depends on the quantity of peptidoglycan cross-link that involves a carboxypeptidase (which corresponds to the SCO4439 gene in the Streptomyces coelicolor genome www.strepdb.com). A Comparison of genome size and number of CDS of 18 Streptomyces genomes with the focus on S Cha CJ, Roe JH (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). Download : Download full-size image; Reconstruction of a genome-scale metabolic model of Streptomyces albus J1074: improved engineering strategies in natural product synthesis. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Two plasmids are similarly available with accession numbers AL589148 and AL645771 . sco1206 encodes RppA which synthesizes 1,3,6,8-tetrahydroxynaphthalene (THN), the intermediate involved in bacterial melanin biosynthesis, by the condensation of five malonyl However, information about such regulatory elements has been limited for Streptomyces genomes.
; Martn, J.F. The genome was divided into 8,645 bins of 1 kb each in a base-by-base moving window, and each of the 51,443 transposon insertions was allocated to a bin. Bentley et al. Summary of Streptomyces coelicolor, Strain A3(2), version 26.0 Tier 2 Curated Database Authors: Georgia Isom 1, Vincent Poon 1, Veronica Armendarez 2, Govind Chandra 2, Mervyn Bibb 2, Gregory L Challis 1, Christophe Corre 1, David Hodgson 1, Jonathan Moore 1 1 University of Warwick, 2 John Innes Centre . 1. 1. Share sensitive information only on official, secure websites. Streptomyces coelicolor and M. tuberculosis are both actinomycetes but have very different lifestyles. Their genomes reveal much similarity at the level of individual gene sequences, and many similar gene clusters. Global comparison showed perceptible higher-order synteny as well, shown as a dot plot in Fig. 2a. strings of text saved by a browser on the user's device. With the advent of genome sequencing technologies, researchers have been working to exploit these data to uncover conserved regulatory elements and link them to a TF. The phages contain DNA ranging in size from 93 to 121 kb and mediate linked transfer of genetic loci at neighboring chromosomal sites sufficiently close to be packaged within the same phage particle. This stage depends on the quantity of peptidoglycan cross-link that involves a carboxypeptidase (which corresponds to the SCO4439 gene in the Streptomyces coelicolor genome www.strepdb.com). A Comparison of genome size and number of CDS of 18 Streptomyces genomes with the focus on S Cha CJ, Roe JH (2006) Identification of genes for mycothiol biosynthesis in Streptomyces coelicolor A3(2). Download : Download full-size image; Reconstruction of a genome-scale metabolic model of Streptomyces albus J1074: improved engineering strategies in natural product synthesis. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Two plasmids are similarly available with accession numbers AL589148 and AL645771 . sco1206 encodes RppA which synthesizes 1,3,6,8-tetrahydroxynaphthalene (THN), the intermediate involved in bacterial melanin biosynthesis, by the condensation of five malonyl However, information about such regulatory elements has been limited for Streptomyces genomes. 
 Furthermore, several genome-scale metabolic models for other biotechnologically relevant Streptomyces strains have been reconstructed since the first S. coelicolor model, iIB711. The fully annotated chromosomal genome is available in the EMBL/GenBank databases with accession number AL645882. The genome of Streptomyces coelicolor, the model organism for this high G+C genus, contains 7825 genes.
Furthermore, several genome-scale metabolic models for other biotechnologically relevant Streptomyces strains have been reconstructed since the first S. coelicolor model, iIB711. The fully annotated chromosomal genome is available in the EMBL/GenBank databases with accession number AL645882. The genome of Streptomyces coelicolor, the model organism for this high G+C genus, contains 7825 genes.  To address this
To address this  We have mapped approximately 10% of the theoretical proteome experimentally using two-dimensional gel electrophoresis and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Author D A Hopwood.
We have mapped approximately 10% of the theoretical proteome experimentally using two-dimensional gel electrophoresis and matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry. Author D A Hopwood. 
 For a bacterium, Streptomyces coelicolor A3 (2) contains a relatively large genome (8.7 Mb) with a complex and adaptive pattern of gene regulation. used an ordered cosmid library to sequence the S. coelicolor genome. Summary: This Pathway/Genome Database The S. coelicolor M145 chromosome is linear and 8.7 Mb in size.
For a bacterium, Streptomyces coelicolor A3 (2) contains a relatively large genome (8.7 Mb) with a complex and adaptive pattern of gene regulation. used an ordered cosmid library to sequence the S. coelicolor genome. Summary: This Pathway/Genome Database The S. coelicolor M145 chromosome is linear and 8.7 Mb in size.  Shah DM (1992) Antifungal proteins from plants. Streptomyces spp. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Abstract Streptomyces coelicolor A3 (2) contains CCC DNA molecules, 2.6 kb in size, with an average copy number of less than one per ten chromosomes. A 1.04 Mb region from the left chromosome end is over-represented in A3 (2)-Stanford, M600 and J1501, but not in CH999. Among them, the genome size of Streptomyces sp. The genome consists of a single linear molecule, and although Ku would be expected to perform end maintenance, none has been observed so far. Streptomyces coelicolor has one linear chromosome and two plasmids, one that is linear and one that is circular. The linear chromosome was sequenced from overlapping clones of the species, most of which were cosmids, that did not contain the two plasmids.
Shah DM (1992) Antifungal proteins from plants. Streptomyces spp. Engineering Streptomyces coelicolor for heterologous expression of secondary metabolite gene clusters. Abstract Streptomyces coelicolor A3 (2) contains CCC DNA molecules, 2.6 kb in size, with an average copy number of less than one per ten chromosomes. A 1.04 Mb region from the left chromosome end is over-represented in A3 (2)-Stanford, M600 and J1501, but not in CH999. Among them, the genome size of Streptomyces sp. The genome consists of a single linear molecule, and although Ku would be expected to perform end maintenance, none has been observed so far. Streptomyces coelicolor has one linear chromosome and two plasmids, one that is linear and one that is circular. The linear chromosome was sequenced from overlapping clones of the species, most of which were cosmids, that did not contain the two plasmids.  RESULTS 4.1.
RESULTS 4.1.  1967 Dec;31(4):373-403. doi: 10.1128/br.31.4.373-403.1967. Complete genome sequence of the model actinomycetes, Huynh QK, Hironaka CM, Levine EB, Smith CE, Borgmeyer JR & Streptomyces coelicolor A3(2).
1967 Dec;31(4):373-403. doi: 10.1128/br.31.4.373-403.1967. Complete genome sequence of the model actinomycetes, Huynh QK, Hironaka CM, Levine EB, Smith CE, Borgmeyer JR & Streptomyces coelicolor A3(2). 
 Genetic analysis and genome structure in Streptomyces coelicolor.
Genetic analysis and genome structure in Streptomyces coelicolor.  Identification and genome-wide comparison between S. parvulus VCCM 22513 and its relatives within the genus Streptomyces.
Identification and genome-wide comparison between S. parvulus VCCM 22513 and its relatives within the genus Streptomyces.  We discovered a correlation between the physical structure of the S.coelicolor genome In most cases (LacI120LacI142 in Fig. Sco7697, a gene encoding a phytase, enzyme able to degrade phytate (myo-inositol 1,2,3,4,5,6-hexakis phosphate), the most abundant phosphorus storing compound in plants is present in the genome of S. coelicolor, a soil born bacteria with a saprophytic lifestyle.
We discovered a correlation between the physical structure of the S.coelicolor genome In most cases (LacI120LacI142 in Fig. Sco7697, a gene encoding a phytase, enzyme able to degrade phytate (myo-inositol 1,2,3,4,5,6-hexakis phosphate), the most abundant phosphorus storing compound in plants is present in the genome of S. coelicolor, a soil born bacteria with a saprophytic lifestyle. 
